讓研究與應用真正連成一線
無論是學術研究或產業需求,艾達特將以整合思維協助您串聯實驗與數據,歡迎與我們聯繫,一起找到最適合的解決方向。
艾達特以定序過程中的 Adaptor(轉接子)為靈感命名,象徵我們在生物與資訊之間承接與轉化的角色。我們致力於從前端樣品建庫到後端數據分析,提供完整而專業的基因體技術服務,成為學術與產業的可信賴夥伴。 在快速變動的時代,我們秉持適存者 (Adaptor) 的精神,協助合作夥伴突破挑戰,開創更多應用與可能。
艾達特致力於建構以客戶需求為核心的基因體技術整合服務平台,推動基因體科技在臺灣及國際的廣泛應用。我們將持續發展:



以本公司所提供之RAD-seq、RNA 基因體技術服務品項為主,提供實驗前討論諮詢。
依照您所取得的生物資訊數據,進行分析前諮詢,確保您所使用的基因體服務符合您的需求。
本公司同時提供您DNA、RNA的客製化分析,可依照您提供的參考文獻進行分析,分析如親緣關係樹建立、遺傳結構分析等。
艾達特提供全方位基因體服務,涵蓋 RAD-seq、RNA 定序、總體基因體(宏基因體)等技術
使用 Illumina 與 Nanopore 平台,從樣本建庫、定序到生物資訊分析,協助推動學術研究、農業改良與產業應用。
限制酶點位核酸定序服務(RAD-seq,Restriction-site Associated DNA Sequencing),以限制酶簡化基因體資訊,提升有效數據比例,可同時處理大量族群樣本,降低研究負擔。RAD-seq 可應用於無參考序列之目標物種的族群遺傳變異、定義個體間遺傳差異、族群親緣地理關係追溯等生物演化研究;同時適用於單核苷酸多態性(SNP,Single Nucleotide Polymorphism)分子標誌建立,可有效應用於農林漁牧等相關產業。

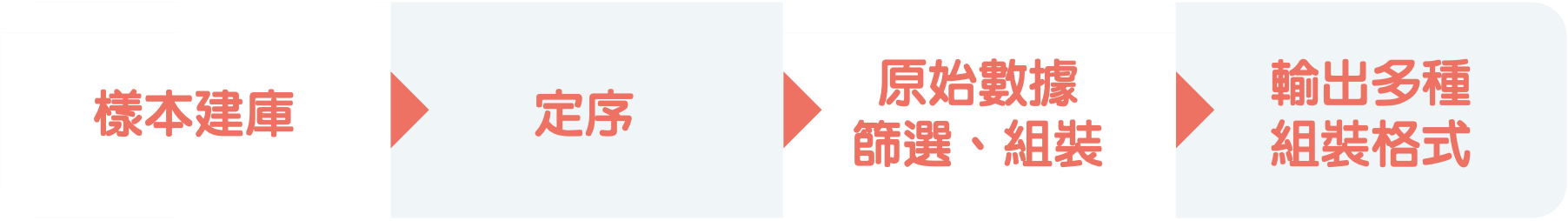
您只需要提供DNA樣品,即可享有樣品建庫、定序,以及生物資訊組裝服務。組裝後的數據可直接供後續各項分析使用,免除研究人員實驗前置作業、數據組裝與處理格式問題所耗時間及設備成本。
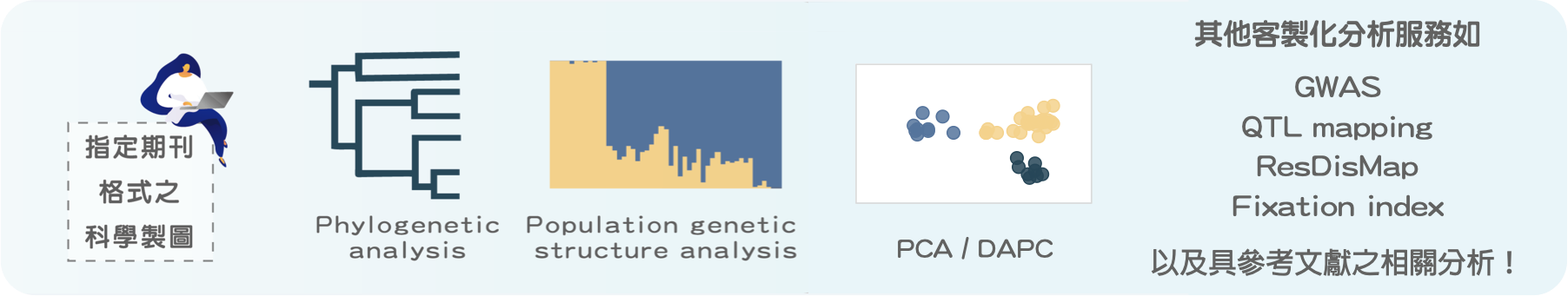
艾達特提供 RAD-seq 組裝後的衍生分析,可輸出符合指定期刊格式之科學圖表。依照需求客製化製圖,強調分析成果亮點,並提供兩次修改與重分析服務,確保分析內容符合研究探討主軸,可提供親緣關係樹分析(Phylogenetic Tree)、族群遺傳結構分析(Population genetic structure)、主成分分析(PCA/DAPC)等,除上列進階分析項目,我們也提供客製化分析服務,可依照您提供的參考文獻進行分析評估,如基因隔離地圖(Dispersal map of resistance)、遺傳分化係數分析(Fixation index analysis)、全基因組關聯分析(GWAS)或數量性狀基因座定位(QTL mapping)等。
在您決定使用艾達特的服務之前,您可以聯繫我們,讓我們為您進行 RAD-seq 的基礎介紹以及研究、應用方向的討論,確保艾達特提供的服務能契合您的期望,同時提供給您樣品測試的服務,透過少量的樣品進行酵素適用性測試,確認您的目標樣品所適合的製備方式。
限制酶點位核酸定序服務(RAD-seq,Restriction-site Associated DNA Sequencing),以限制酶簡化基因體資訊,提升有效數據比例,可同時處理大量族群樣本,降低研究負擔。RAD-seq 可應用於無參考序列之目標物種的族群遺傳變異、定義個體間遺傳差異、族群親緣地理關係追溯等生物演化研究;同時適用於單核苷酸多態性(SNP,Single Nucleotide Polymorphism)分子標誌建立,可有效應用於農林漁牧等相關產業。

您只需要提供DNA樣品,即可享有樣品建庫、定序,以及生物資訊組裝服務。組裝後的數據可直接供後續各項分析使用,免除研究人員實驗前置作業、數據組裝與處理格式問題所耗時間及設備成本。

艾達特提供 RAD-seq 組裝後的衍生分析,可輸出符合指定期刊格式之科學圖表。依照需求客製化製圖,強調分析成果亮點,並提供兩次修改與重分析服務,確保分析內容符合研究探討主軸,可提供親緣關係樹分析(Phylogenetic Tree)、族群遺傳結構分析(Population genetic structure)、主成分分析(PCA/DAPC)等,除上列進階分析項目,我們也提供客製化分析服務,可依照您提供的參考文獻進行分析評估,如基因隔離地圖(Dispersal map of resistance)、遺傳分化係數分析(Fixation index analysis)、全基因組關聯分析(GWAS)或數量性狀基因座定位(QTL mapping)等。

在您決定使用艾達特的服務之前,您可以聯繫我們,讓我們為您進行 RAD-seq 的基礎介紹以及研究、應用方向的討論,確保艾達特提供的服務能契合您的期望,同時提供給您樣品測試的服務,透過少量的樣品進行酵素適用性測試,確認您的目標樣品所適合的製備方式。


Adaptor Genomic Sciences 的新聞




無論是學術研究或產業需求,艾達特將以整合思維協助您串聯實驗與數據,歡迎與我們聯繫,一起找到最適合的解決方向。